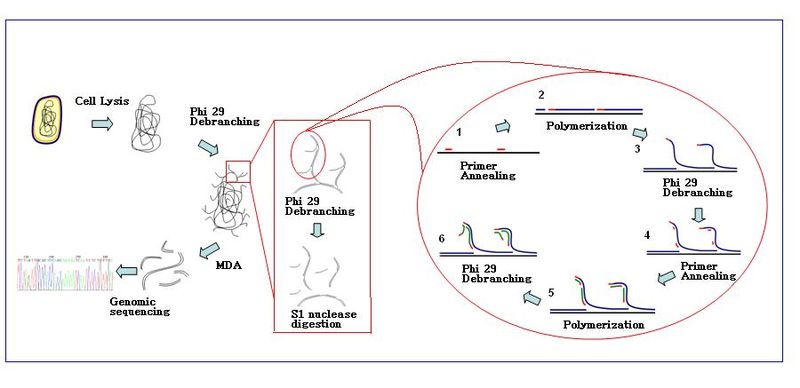
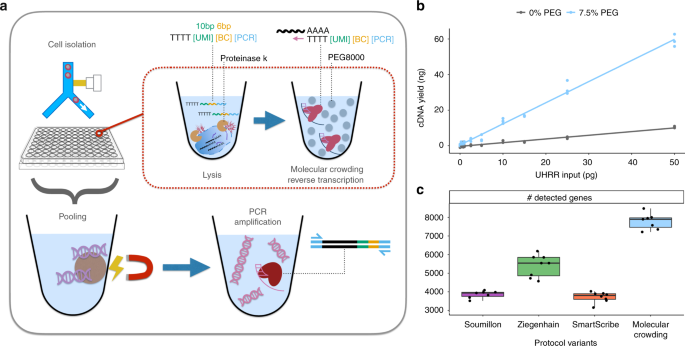
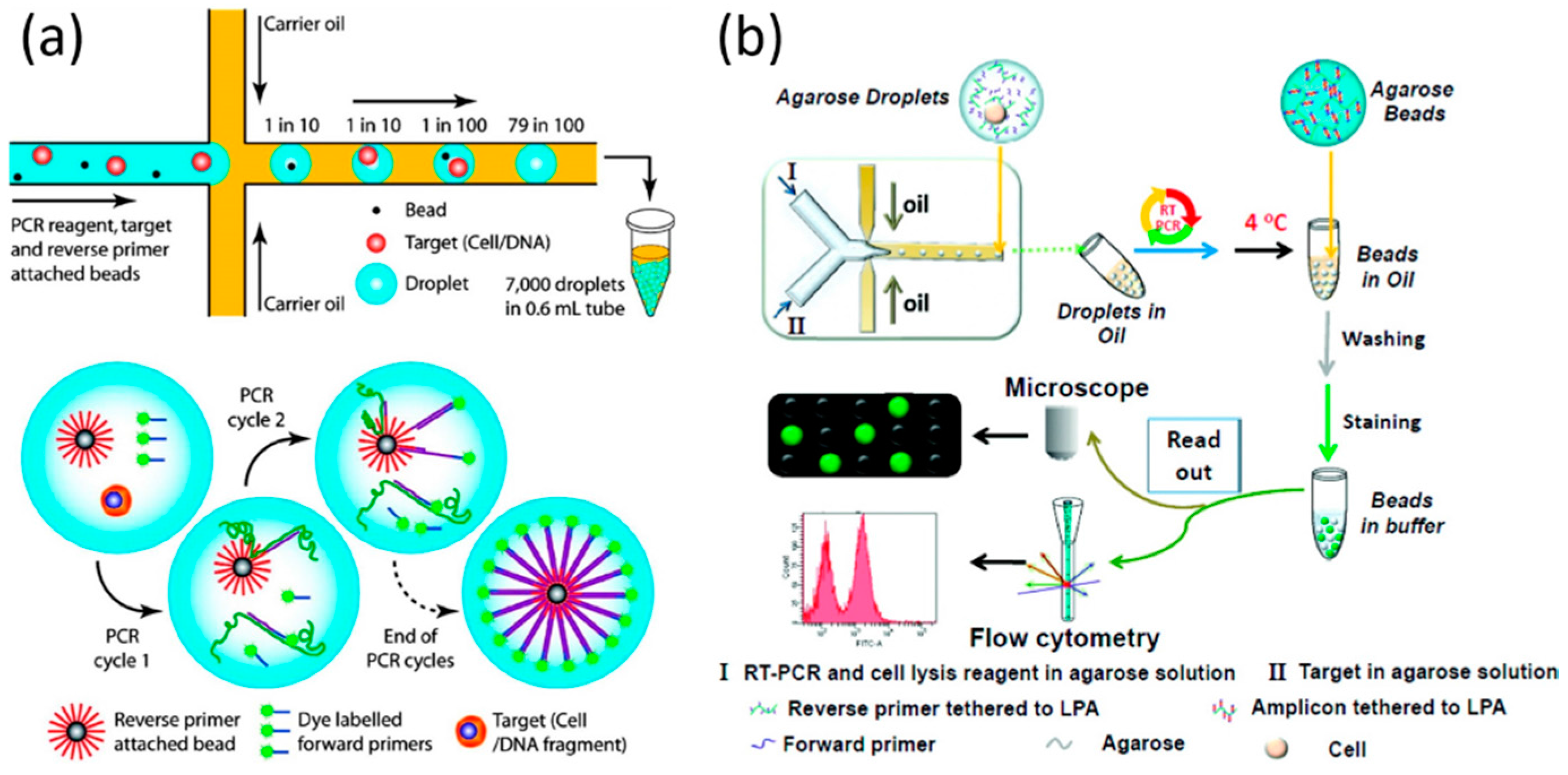
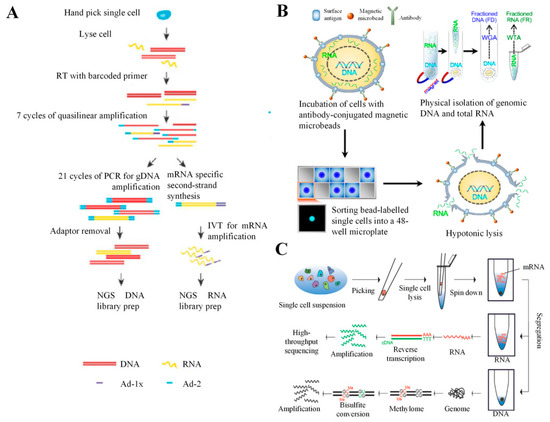
Multiple displacement amplification mda is a widely used technique enabling amplifying femtograms of dna from bacterium to micrograms for the use of sequencing.
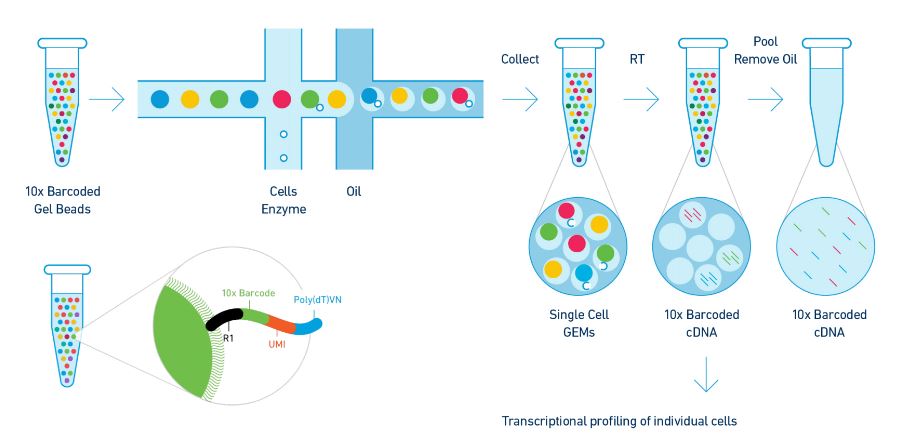
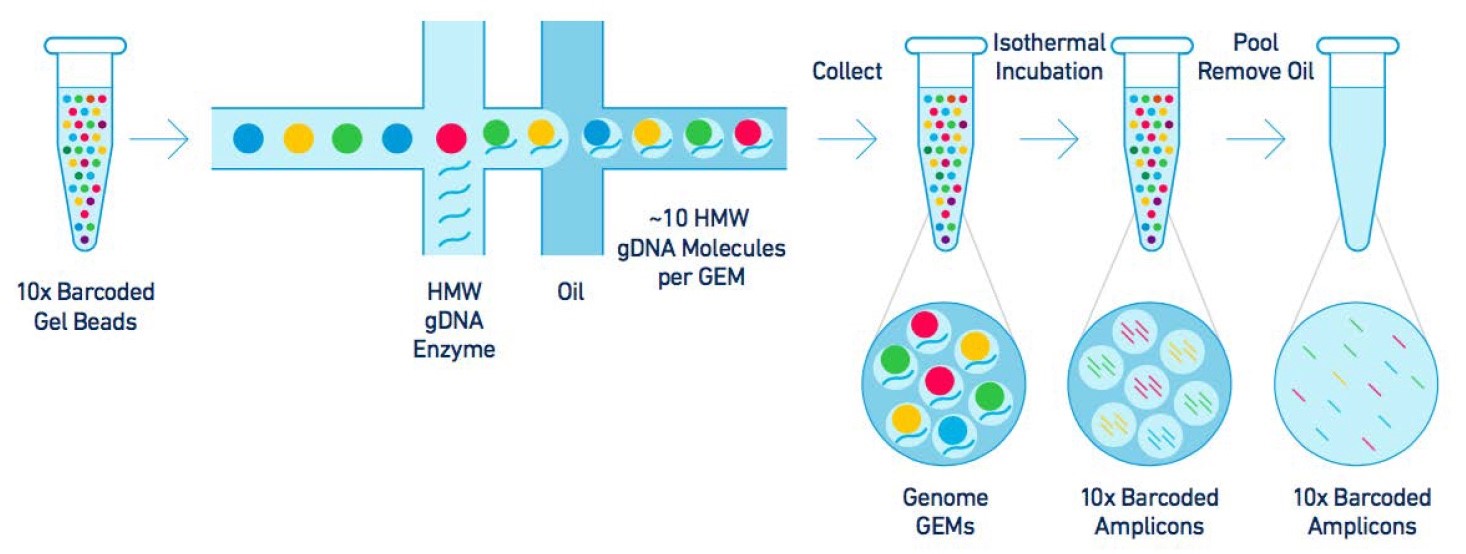
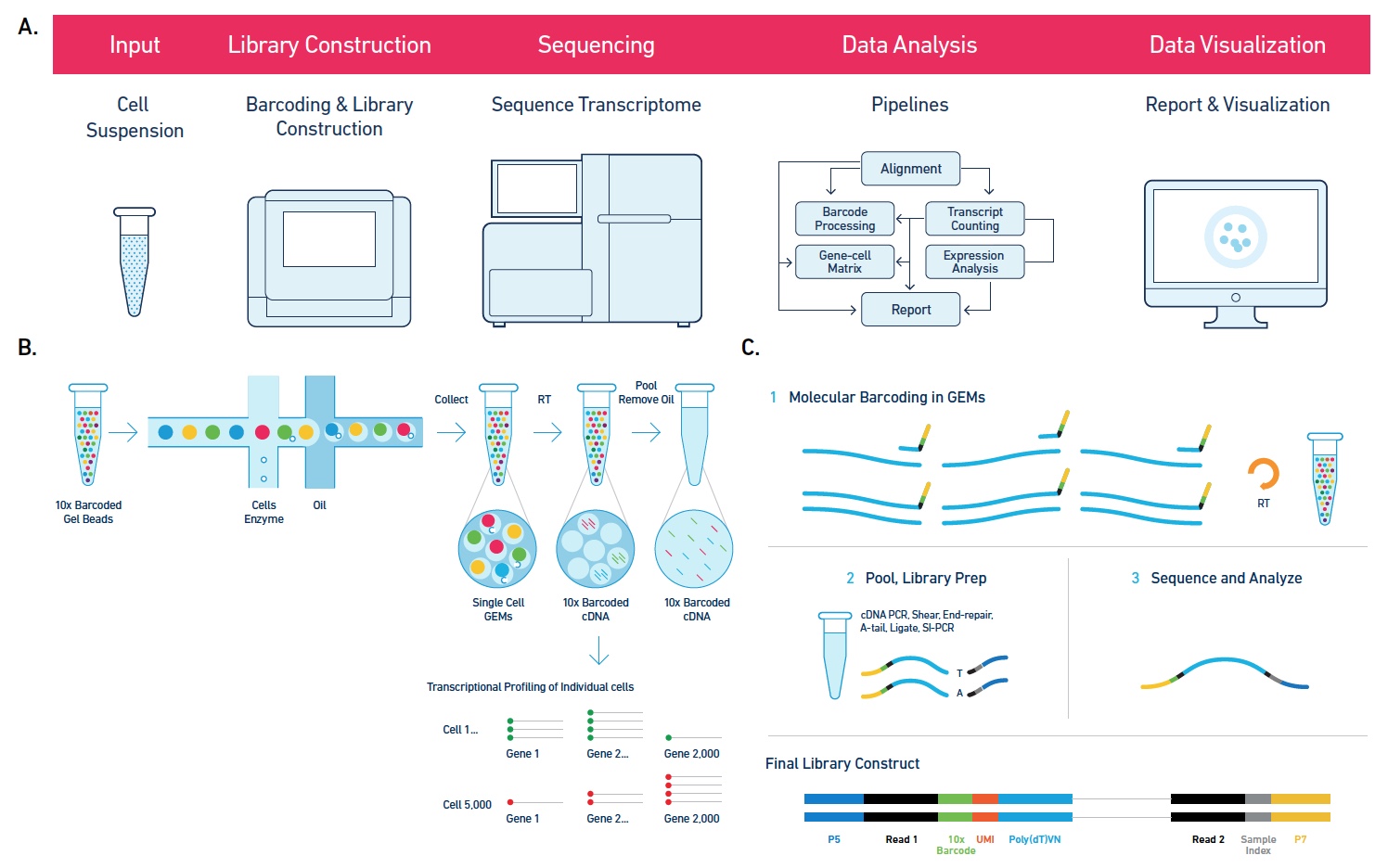
Single cell genome amplification.
1 the kit includes a robust optimized cell lysis protocol that is incorporated into the.
Here we report an improved single cell wga method linear amplification via trans.
The key parameters to characterize the performance of these methods are defined including genome coverage uniformity.
A list of more than 100 different single cell omics methods have been published.
Single cell genomics is important for biology and medicine.
1 pg of e coli dna has been amplified with either the repli g single cell kit or a kit based on malbac technology library prep and miseq sequencing similar data analysis on clc workbench.
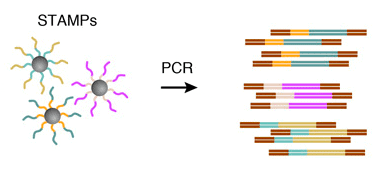
Genomeplex single cell whole genome amplification kit utilizes a proprietary technology based on random fragmentation of genomic dna and conversion of the resulting small fragments to pcr amplifiable library molecules flanked by universal priming sites.
Background whole genome amplification wga is currently a prerequisite for single cell whole genome or exome sequencing.
At very low levels of input dna successful amplification relies on reagent quality.
Single cell genomiphi dna amplification kit has been optimized to wholly amplify genomic dna from as little as a single cell generating micrograms of high quality dna for use in downstream applications.
Wga is achieved by pcr amplification of the library molecules using universal.
Single cell whole genome amplification and sequencing.
Single cell genomiphi has been developed with this in mind.
Methodology and applications annu rev genomics hum genet.
Depending on the method used the rate of artifact formation allelic dropout and sequence coverage over the genome may differ significantly.
Authors lei huang 1 fei ma alec chapman sijia lu xiaoliang sunney xie.
Results the largest difference between the evaluated protocols was observed when analyzing the target coverage and read depth.
Random primers and dna polymerase from bacteriophage phi29.
Consequently accurate whole genome amplification wga of single cell dna is required for reliable genetic analysis e g ngs.
E coli as reference genome is an ideal model system to identify amplification bias introduced via technology used.
However current whole genome amplification wga methods are limited by low accuracy of copy number variation cnv detection and low amplification fidelity.
Whole genome amplification from a single cell is now possible with our optimized genomeplex single cell whole genome amplification kit the single cell procedure differs very little from the previously described genomeplex system but for three procedural changes.
We present a survey of single cell whole genome amplification wga methods including degenerate oligonucleotide primed polymerase chain reaction dop pcr multiple displacement amplification mda and multiple annealing and looping based amplification cycles malbac.
In contrast a deep analysis of the genome currently requires a few hundred nanograms up to micrograms of genomic dna for library formation necessary for ngs sequencing or labeling protocols e g microarrays.